GUEST LECTURES
Euan James
The James Hutton Institute, Scotland UK
Use of Transmission Electron Microscopy (TEM) in studies on plant-microbe interactions
TEM involves the transmission of an electron beam through a sample so that detail can be discerned of areas that are more or less opaque to the electrons. It requires very thin samples e.g. sections (c. 70 nm), which are cut using an ultramicrotome. The tissues in the sections are differentially stained using solutions of heavy metal salts (e.g. those of lead and uranium) which deflect the electron beam. TEM is generally used for the analysis of internal structure of cells, and together with immunogold labeling (IGL) specific sites (“antigens”) on TEM sections can be identified by incubating the section in an antibody which has been raised against the specific antigen. The primary antibody binds specifically to the corresponding antigen in the sample, and these binding sites can then be visualized under the TEM after incubation of the sample in a “secondary antibody” which has been conjugated to a gold particle (usually ranging in diameter from 5 – 20 nm). The IGL can be quantified. We routinely use TEM-IGL in tandem with light microscopy (conventional and fluorescence) to investigate interactions between beneficial bacteria and their host plants at the sub-cellular level; these include Nitrogen (N)-fixing bacterial symbionts residing in the cells in legume root and stem nodules, and N-fixing endophytic bacteria in non-legumes, such as cereals. A recent example of this approach is in studies on the evolution of a “stable” nodulating symbiosis with rhizobia. Recent evidence suggesting a single origin of nodulation followed by massive parallel evolutionary losses raises questions about why a few lineages in the N2-fixing clade retained nodulation and diversified as stable nodulators while most did not. A nodule anatomy type that is predominant across the legume subfamily Caesalpinioideae, in which there have been extensive losses of nodulation, are those wherein the rhizobial symbiont is not symplastically internalized in the host cell cytoplasm within membrane-bound symbiosomes, but are instead retained within the apoplast surrounded by a cell wall in so-called “fixation threads” (FTs). A statistical analysis applied to an updated phylogeny of the Caesalpinioideae indicates that symbiosis in FT-type nodule is less stable and more prone to losses owing to its less tightly compartmentalized integration of the two partners. TEM has demonstrated that the FTs are, in fact, comprised of a “sandwich” in which the cell wall material is surmounted by a symbiosome membrane; we suggest that the FT cell wall confers an additional protective layer to the rhizobia should they be perceived by the host as “pathogenic” in this less intimate association.
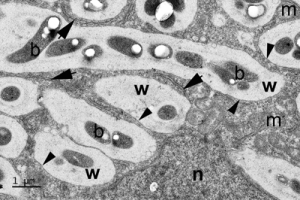
Upper panel, section of a root nodule from the West African legume tree Erythrophleum suaveolens; rhizobial bacteroids (b) are contained within newly-formed FTs packed into a new N-fixing host plant cell.
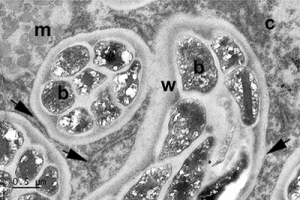
Lower panel, section of a root nodule from the Brazilian legume tree Moldenhawera floribunda with its N-fixing bacteroids (b) enclosed within mature FTs.
In both cases, note the thick wall (w) of the FTs, the membranes associated with them (arrow heads), and the strands of endoplasmic reticulum from which the membranes are derived (arrows). c = cytoplasm, m = mitochondrion, n = nucleus.
Reference
Sprent et al. (2017) New Phytologist 215: 40-56.
Hanns-Heinz Kassemeyer
„The plant surface as a preformed barrier and contact zone in the host-pathogen interaction“
Staatliches Weinbauinstitut Freiburg: www.wbi-bw.de
Ovečka Miroslav
Centre of the Region Haná for Biotechnological and Agricultural Research, Faculty of Science, Palacký University Olomouc, Šlechtitelů 27, 783 71 Olomouc, Czech Republic
http://cr-hana.upol.cz/cellbiol/
@Samaj_lab
Light sheet microscopy in plant developmental studies and plant-microbe interaction studies
Live cell imaging in plants is prone to several limitations, like specific optical properties of plant cell surfaces, presence of vacuoles, as well as environmental requirements for undisturbed plant development and growth during imaging. Subcellular architecture, organization and dynamics of most organelles in cells of living plants can be monitored at the nanoscale using super-resolution microscopy methods, such as structured illumination microscopy (SIM), while long term monitoring of the dynamic cellular changes in a developmental context can be monitored in whole plant organs by light sheet fluorescence microscopy (LSFM). We developed reliable and broadly applicable protocols allowing short term bioimaging of plant cells using SIM (Komis et al. 2015) and long term developmental imaging of plants using LSFM (Ovečka et al. 2015).
We applied these advanced microscopy methods for clarification of diverse scientific questions, such as monitoring of microtubule and actin dynamics during plant morphogenesis, signalling by mitogen-activated protein kinase during cell division, nuclear changes in diverse root developmental zones and tissues, contribution of secretory vesicles to the oscillatory growth of root hairs, or dynamic behaviour and interactions of endosomes in growing root hairs. Although most of these studies were performed in the model plant Arabidopsis, we successfully established LSFM imaging also in crops such as alfalfa, tobacco and barley. Taking together, time lapse super-resolution imaging of plant cells as well as developmental imaging of whole plants under gentle environmental conditions significantly advanced our understanding of plant growth and development.
This work was supported by ERDF project ‘Plants as a tool for sustainable global development’ (CZ.02.1.01/0.0/0.0/16_019/0000827).
References
Komis et al., Nat. Protoc. 10 (2015) 1248-1263.
Ovečka et al., Nat. Protoc. 10 (2015) 1234-1247.